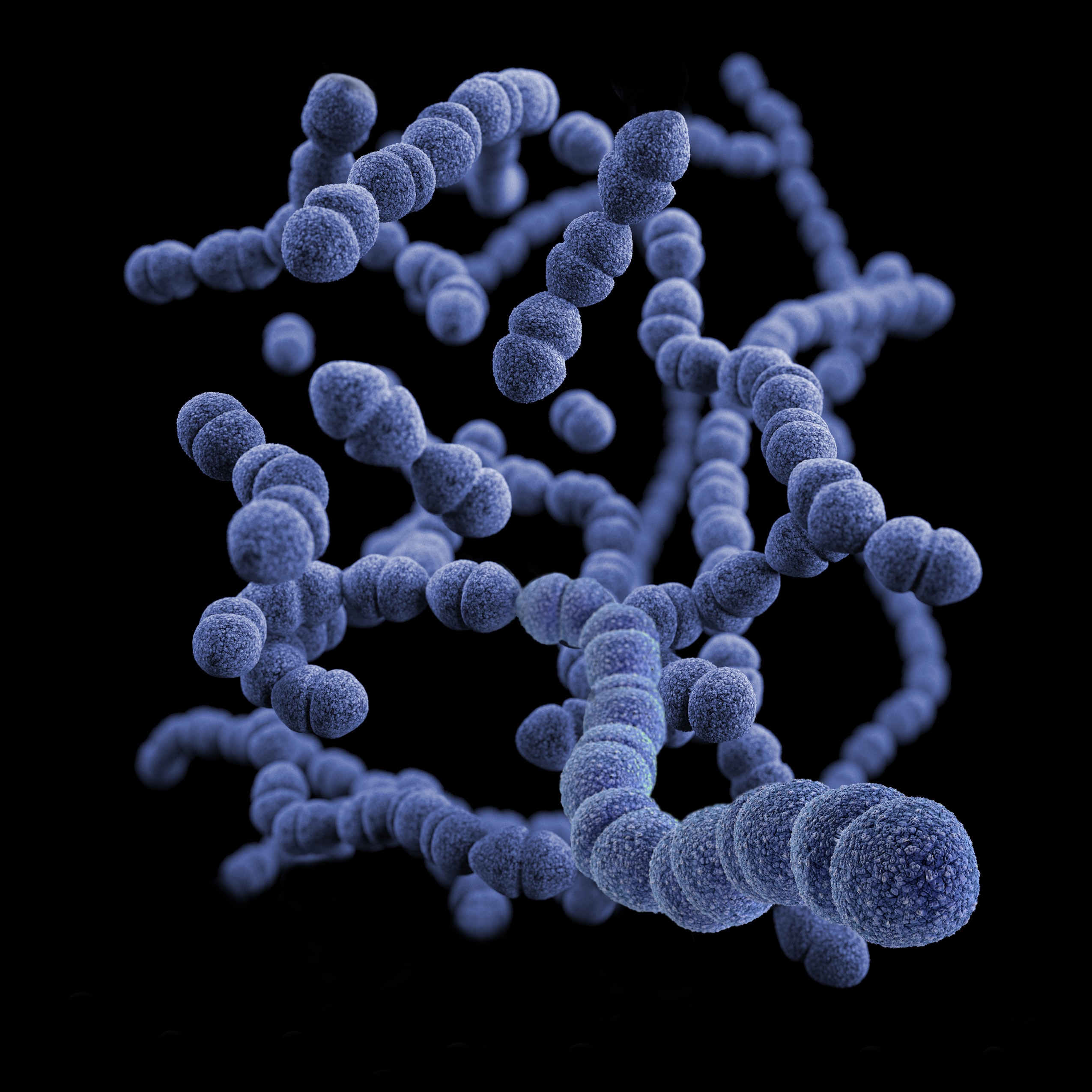
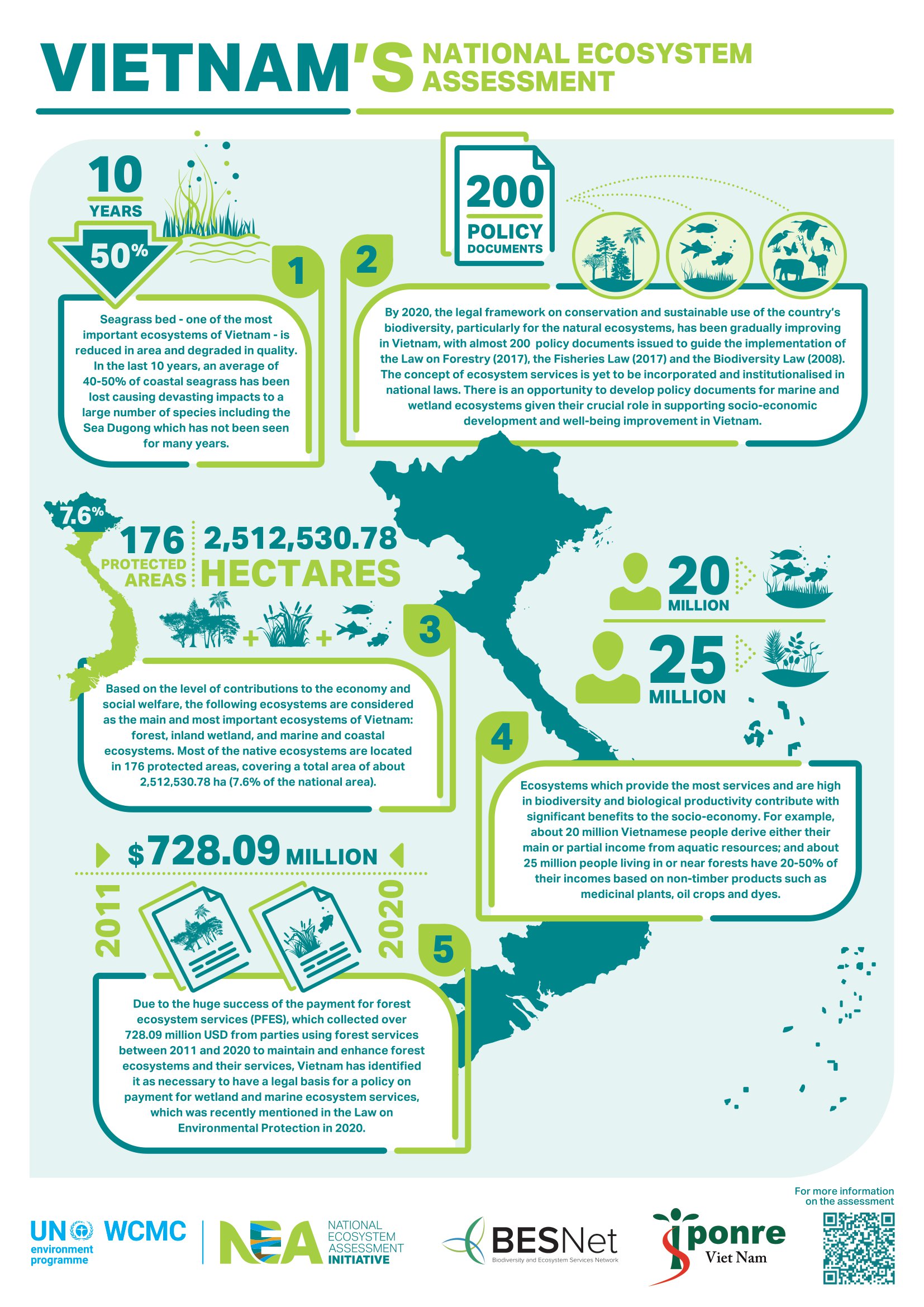
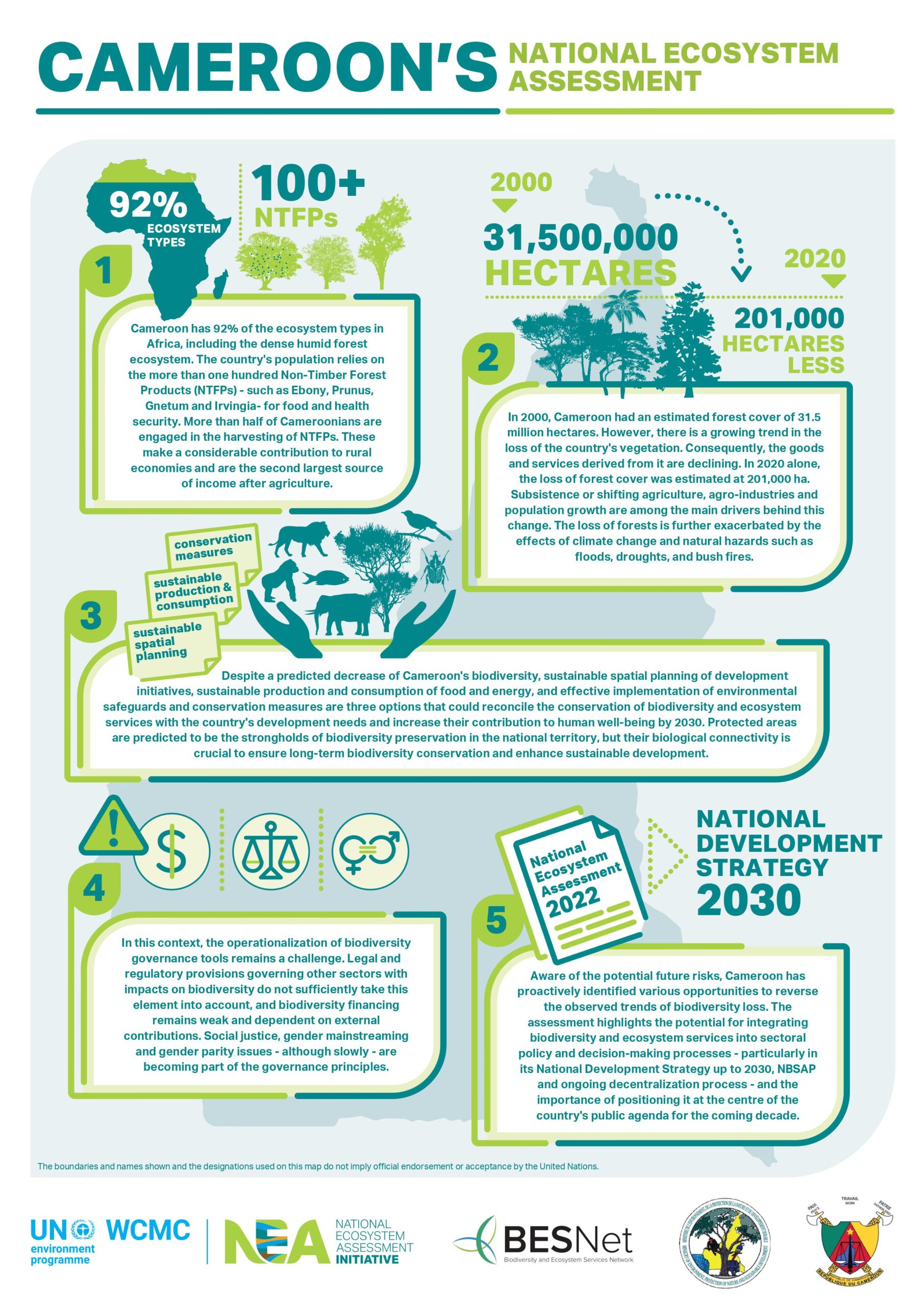
High-throughput high-density genotyping arrays continue to be a fast, accurate, and cost-effective method for genotyping thousands of polymorphisms in high numbers of individuals. Here, we have developed a new high-density SNP genotyping array (103,270 SNPs) for honey bees, one of the most ecologically and economically important pollinators worldwide. SNPs were detected by conducting whole-genome resequencing of 61 honey bee drones (haploid males) from throughout Europe. The selection of SNPs for the chip was done in multiple steps using several criteria. The majority of SNPs were selected based on their location within known candidate regions or genes underlying a range of honey bee traits, including hygienic behavior against pathogens, foraging, and subspecies. Additionally, markers from a GWAS of hygienic behavior against the major honey bee parasite Varroa destructor were brought over. The chip also includes SNPs associated with each of three major breeding objectives— honey yield, gentleness, and Varroa resistance. We validated the chip and make recommendations for its use by determining error rates in repeat genotyping, examining the genotyping performance of different tissues, and by testing how well different sample types represent the queen’s genotype. The latter is a key test because it is highly beneficial to be able to determine the queen’s genotype by nonlethal means. The array is now publicly available and we suggest it will be a useful tool in genomic selection and honey bee breeding, as well as for GWAS of different traits, and for population genomics, adaptation, and conservation questions.
Tool for genomic selection and breeding to evolutionary adaptation: Development of a 100K single nucleotide polymorphism array for the honey bee
Year: 2020